E1.2: Structure-Function Analysis of Trojan Peptides Used for Transporting Nanomaterials into Cells
Subproject Leader: Anne Ulrich
Institut für Organische Chemie, KIT
Contributing Scientists:
Present: Nico Heidenreich, Julia Misiewicz, Sabine Reisser
Past: Susanne Fanghänel, Silke Hoffmann, Katja Koch, Volodymyr Kubyshkin, Ronald Lochte, Jan Roll, Serge Ruden
Peptides as “Magic Bullets”
More than 100 years ago the immunologist Paul Ehrlich conceived the idea of using molecules as “magic bullets” to target and destroy dangerous pathogens. Today, certain peptides have been discovered which indeed act as genuine molecular bullets by physically penetrating the membrane barrier of cells. Such peptides can serve two different kinds of biological functions. Cell penetrating peptides pass through membranes in a non-leaky fashion and carry cargo along, while antimicrobial peptides damage the protective membrane barrier of bacteria and kill the pathogen. Both mechanisms are useful for nano-biological applications, depending on the cell type against which the peptides are targeted and the kind of membrane-perturbation they induce. These peptides and their interactions with nanoparticles are being studied in this sub-project E1.2, using diverse biophysical methods with a special focus on solid-state NMR and circular dichroism spectroscopy, and using MD simulations in sub-project E1.6.
Structure Analysis of Membrane-Bound Peptides
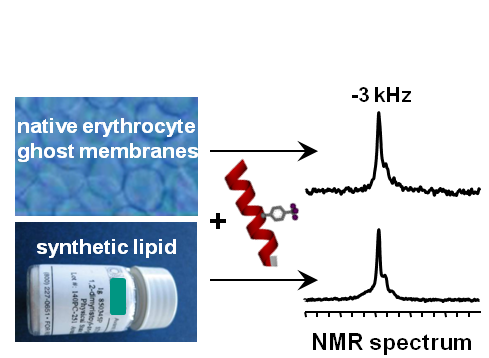
To understand the molecular mode of action of a membrane-active peptide, it is essential to know its 3D structure, its dynamic behaviour, its self-assemble into specific oligomers, or its tendency to aggregate non-productively. Solid-state NMR is ideally suited to characterize selectively labeled peptides using 2H-/13C-/15N-NMR, and to monitor the response of the lipid bilayer using 31P-NMR. To boost the intrinsically low sensitivity of conventional NMR by some orders of magnitude, a highly sensitive and background-free 19F-labeling scheme for peptides has been developed. Specific amino acids with CF3-reporter groups yield unique orientational NMR-parameters for the 3D structure calculation. Numerous representative peptides have thus been characterized in lipid model membranes, and distinct membrane-permeabilization mechanisms could be described [1-4]. Recently, a 19F-labeled peptide has even be observed and analyzed in native biomembranes, harvested from human erythrocytes or from bacterial protoplasts, which paves the way for in-cell NMR [5]. Additionally, qualitative information about the peptide conformation and orientation in membranes can be obtained by circular dichroism (CD), using the same oriented samples as for solid-state NMR. A new VUV-CD beamline, acquired from the Daresbury Laboratory, has been installed in 2010 at the ANKA synchrotron in Karlsruhe.
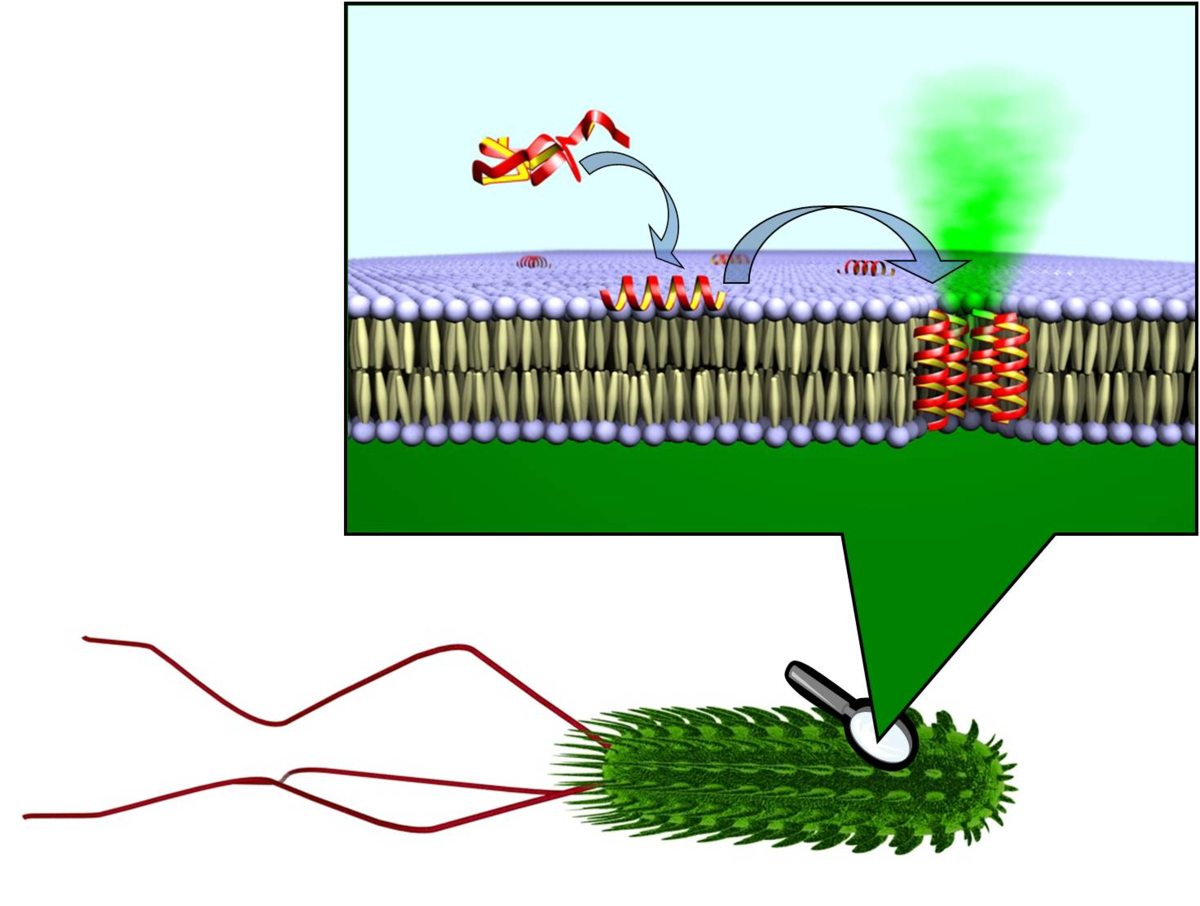
Cell Penetrating Peptides: Deliver Cargo to a Specific Target
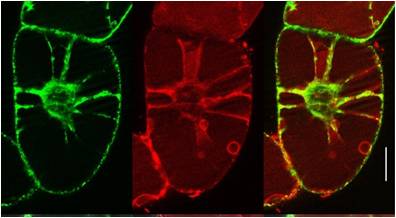
Amongst many established cell penetrating peptides investigated, such as HIV-TAT or transportan, the most promising candidate for delivery into plant cells was found to be BP100 (see sub-project E1.4). It was originally optimized by medium-throughput screening as an antimicrobial agent against plant pathogens. The very short sequence is attractive for coupling it to other peptidic moieties. When connected to the actin-binding sequence Lifeact, the BP100 construct could thus be selectively targeted to actin filaments in tobacco cells. It allowed the cytoskeleton to be visualized, as demonstrated by the Nick group. Solid-state 19F-NMR showed that the very short BP100 inserts upright into membranes with an a-helical structure and assembles into an extended oligomeric complex.
Antimicrobial Peptides: Search and Destroy the Enemy
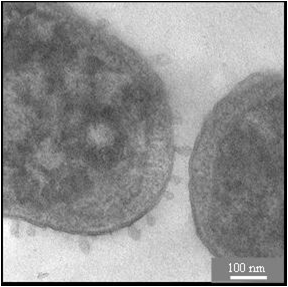
Antimicrobial peptides share many physicochemical features with cell penetrating sequences, such as the synergistic pair PGLa and Magainin in the skin of frogs. Solid state 19F-NMR analysis suggests that these a-helices can assemble into transient tranmembrane pores. Temporin A is another antimicrobial peptide from frog skins, but due to its much shorter sequence it cannot span the lipid bilayer. Solid-state 19F-NMR suggests a similar structure and alignment as BP100, but a much higher mobility implies a monomer that can undergo flip-flop. To increase the efficiency of antimicrobial peptides, they can be tethered to nanoparticles to be exposed in multiple copies. Specifically in the case of silver nanoparticles, a synergistic effect was observed even by simply mixing them with certain antimicrobial peptides. The membrane-perturbing effects on cells are visualized by electron microscopy in collaboration the Gerthsen group.
References
|
[1] |
S. Afonin, S.L. Grage, M. Ieronimo, P. Wadhwani, and A.S. Ulrich, J. Am. Chem. Soc. 130, 16512 (2008) |
|
[2] |
P. Wadhwani, J. Bürck, E. Strandberg, C. Mink, S. Afonin, M. Ieronimo, and A.S. Ulrich, J. Am. Chem. Soc. 130, 16515 (2008) |
|
[3] |
S. Afonin, U.H.N. Dürr, P. Wadhwani, J. Salgado, and A.S. Ulrich, Topics Curr. Chem. 273, 139 (2008) |
|
[4] |
D. Maisch, P. Wadhwani, S. Afonin, C. Böttcher, B. Koksch, and A.S. Ulrich, J. Am. Chem. Soc. 131, 15596 (2009) |
|
[5] |
M. Ieronimo, S. Afonin, K. Koch, M. Berditsch, P. Wadhwani, and A.S. Ulrich, J. Am. Chem. Soc. 132, 8822 (2010) |
List of Publications 2006-2011 as PDF
Subproject Report 2006-2010 as PDF