C5.4: Protein-Assisted Assembly of Superlattices
Subproject Leader: Holger Puchta
Contributing Scientists:
Present: Anna Barth, Manfred Focke, Daniela Kobbe
Past: Markus Bauknecht, Sandra Blanck, Verena Eschbach, Martin Meng
Proteins as Toolbox for Modifying and Assembling DNA Modules
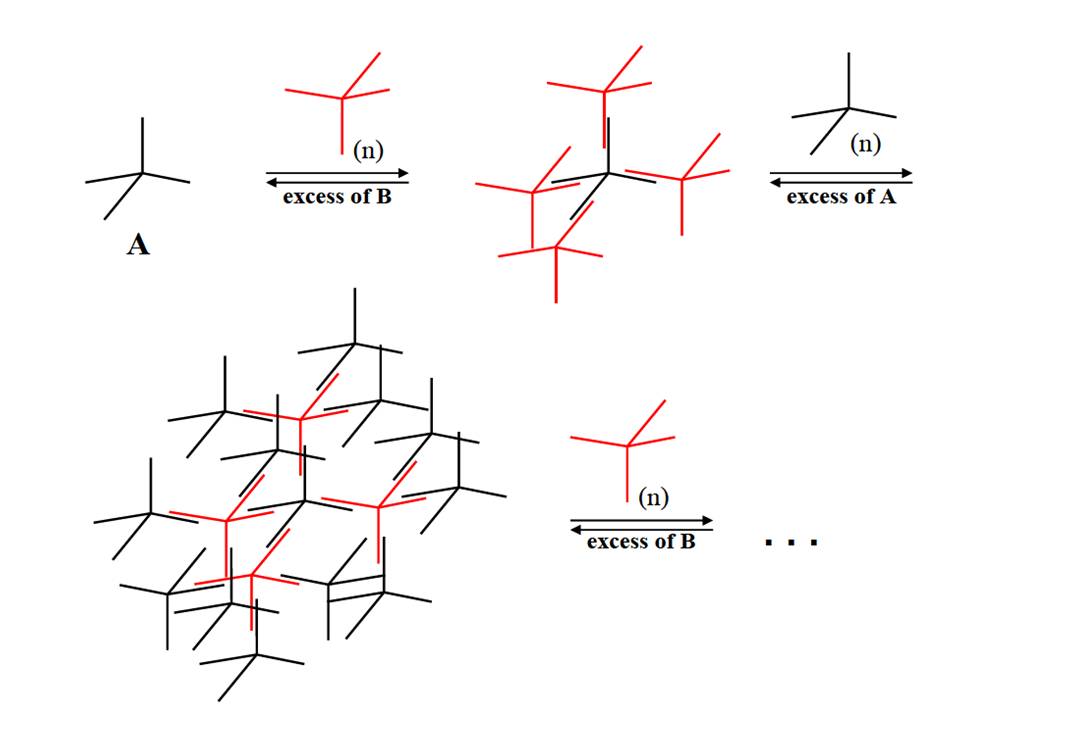
In subproject C5.4: "Protein assisted assembly of superlattices" DNA hybrid molecules are used to build up ordered DNA structures. Such DNA hybrid molecules are made up of a tetraphenylmethane (TPM) core, synthesized by our cooperation partner project C5.2 (AG Stefan Bräse) linked to four non-complementary oligonucleotide arms in which the 3’-ends are covalently attached to the phenyl moieties and the free 5’ends are orientated to the exterior. The step-wise synthesis of the oligonucleotide arms was done by in the course of the C5.3 project (AG Clemens Richert). Because the length of the oligonucleotide arms by chemical means is restricted to 6-8 bases, we elongated the hybrid molecule with the aid of proteins. This was accomplished by phosphorylating the 5’ end with polynucleotide kinase, annealing the hybrid TPM with a splint oligonucleotide and an elongating oligonucleotide and closing the nick between the TPM oligonucleotide and the elongation oligonucleotide by the enzyme T4 DNA ligase. This technique enables us to produce TPM molecules with oligonucleotide arms of different length and sequence. The further strategy was to synthesize two molecular species of elongated TPMs called A and B in which the oligonucleotide arms are complementary to the respective counterpart. The theoretical calculations and modeling of the expected supra-molecular structures were done by the group of Wolfgang Wenzel (C5.1).
Assembling DNA Hybrid Molecules
For the assembly of higher ordered DNA structures (alternating shells of the TPM A and B) it is essential to control the speed and precision of the assembly process. The first shell (AB4 structure) could be annealed by conventional methods. This includes the incubation in tetraethylammonium chloride (TEAC) und the purification of the AB4 structure by preparative gel electrophoresis. Further shell layers need a more sophisticated annealing concept. This can be done with proteins/enzymes. In the course of our research on DNA modifying enzymes we identified, cloned, expressed and purified helicases (SRS2 [1], RECQ3 [2]) that exhibit beside their strand displacement also a strand annealing activity. The manipulation of the balance between displacement and annealing activity can be an important tool to influence the assembly process of DNA building blocks. Further research is at the moment done on single strand binding proteins such as the protein RPA. The sequestering of single stranded regions in a DNA module might be very helpful to suppress unwanted annealing events.
References
| [1] | S. Blanck, D. Kobbe, F. Hartung, K. Fengler, M. Focke, and H. Puchta , Nucl. Acids Res. 37, 7163 (2009) |
| [2] | D. Kobbe, S. Blanck, M. Focke, H. Puchta, Plant Physiol. 151, 1658 (2009) |
List of Publications 2006-2011 as PDF
Subproject Report 2006-2010 as PDF